| Version 1 (modified by , 15 years ago) (diff) |
|---|
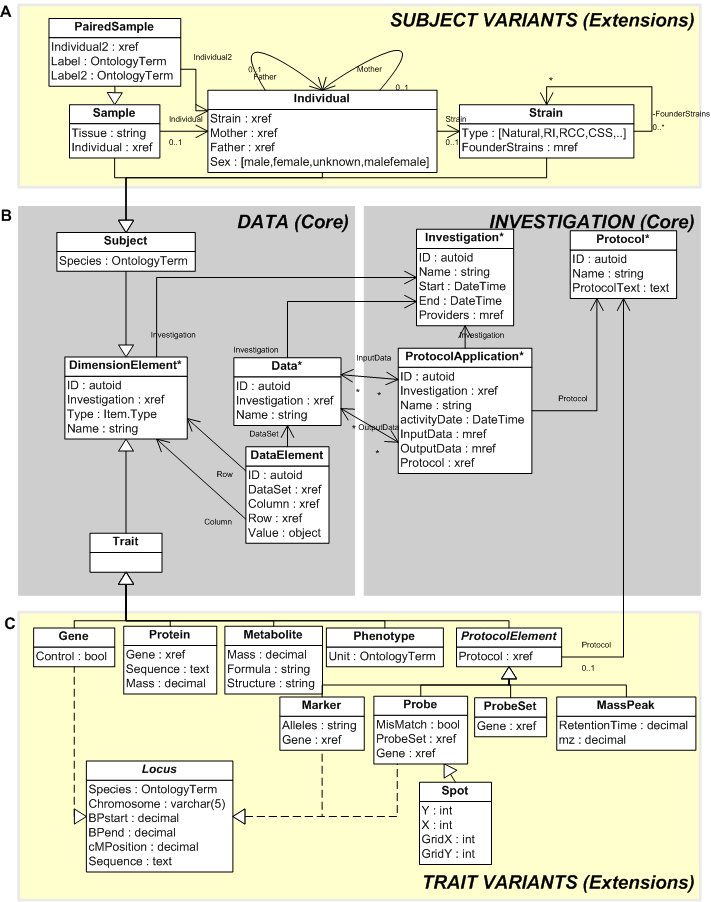
XGAP Object Model
XGAP-OM is the conceptual model behind the XGAP platform. It is designed to provide uniform model for a wide variety of organisms, experimental designs, and biomolecular profiling technologies:
- A core for the raw, in process and result data using only four core data types Trait, Subject, Data and DataElement.
- A core to for experimental design annotations reusing FuGE data types Investigation, Protocols and ProtocolApplications, OntologyTerms, etc.
- Consistently annotate Traits and Subjects using standardized extensions of Trait (e.g. Probe, Marker) and Subject (e.g. Individual, Strain).
- Consistently extend XGAP for new types of annotations by adding more types of Strain and Subject (e.g. add 'MassPeak' as a new Trait to annotate 'retentiontime' and 'mz')
Specifications
- Overview of XGAP-OM v1.0 in UML (xgap_umldiagram_v1_0.pdf)
- Informal description of each XGAP-OM v1.0 data entity (generated docs)
- Formal definition of XGAP-OM v1.2 in MOLGENIS* language(xgap_db.xml)
- Formal definition of FuGE-OM in MOLGENIS* language (fuge_db.xml)
*We use MOLGENIS to auto-generate software from the XGAP-OM. To ease integration with other research domain, XGAP has been structured as full extension of functional genomics standard FuGE . When generating software from the data model one also needs the FuGE definition. See XgapCustomization.
Attachments (6)
- xgap_umldiagram_v1_0.pdf (21.1 KB) - added by 16 years ago.
- xgap-om.gif (62.2 KB) - added by 16 years ago.
- fuge_db.xml (24.7 KB) - added by 16 years ago.
- fuge_db.2.xml (24.7 KB) - added by 16 years ago.
- xgap_db_v1_0.xml (15.0 KB) - added by 16 years ago.
- xgap_ui.xml (1.7 KB) - added by 16 years ago.
Download all attachments as: .zip